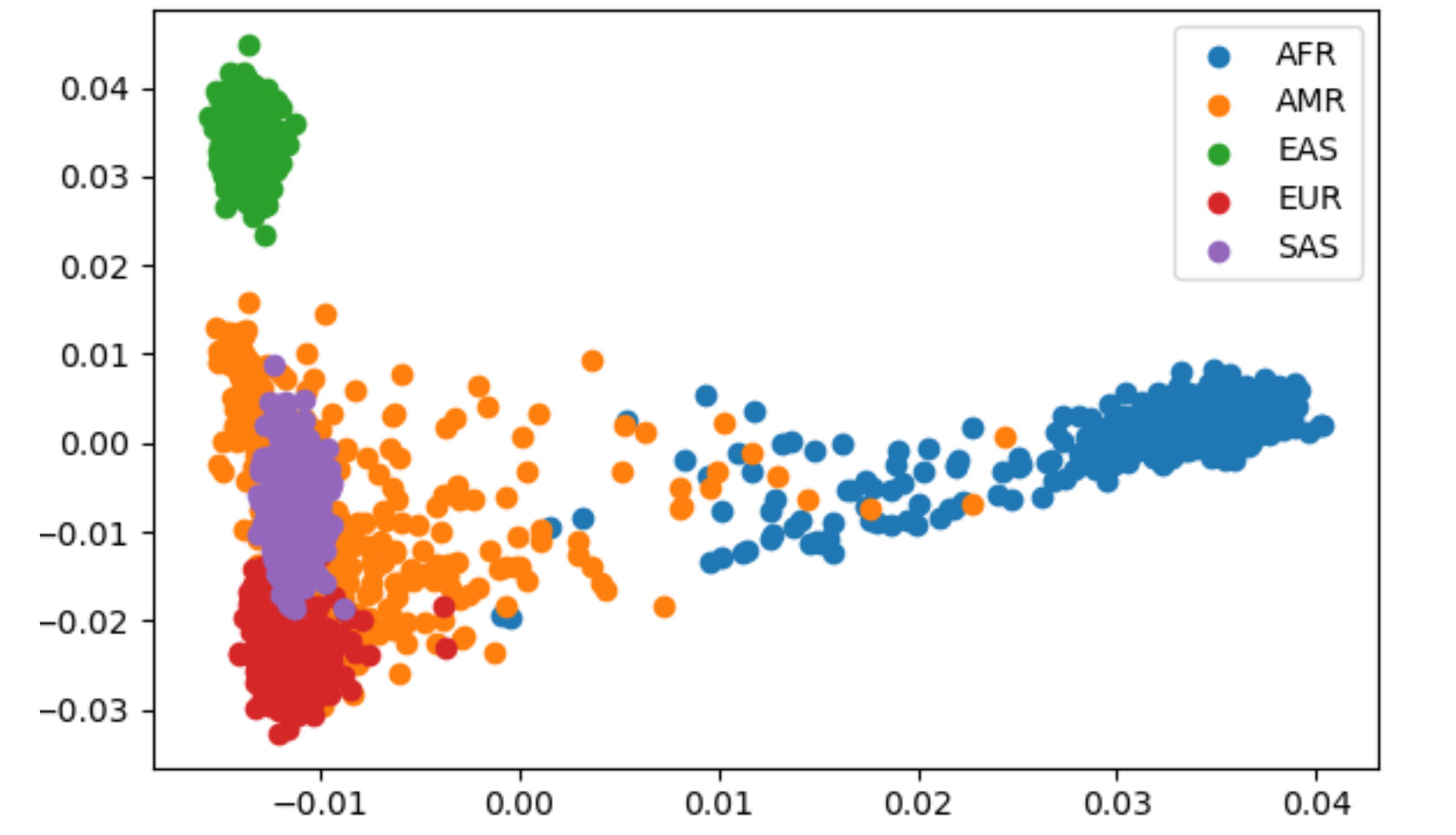
Teaching
Human Genome Variation Lab
In 2020, I was awarded a $4000 Instructional Enhancement Grant from the Johns Hopkins Center for Teaching Excellence & Innovation.
With help from Rajiv McCoy and Kate Weaver, I used this grant to develop a semester's worth of open-source modules for introducing undergraduates to human genetics. These modules are used to teach the Human Genome Variation Lab course at Johns Hopkins (AS.020.319).
The course materials, available as an online textbook and "starter code" R Markdown files, adapt public datasets to:
- Introduce key concepts from human genetics and evolution
- Teach the basics of data analysis in R
- Encourage hands-on learning through built-in coding exercises and homework
I am happy to share teaching materials from our undergrad computational course on human genetic variation for other students and instructors to use and adapt. This was developed by @smyan__ with help from @KjweavI and me, supported by @JHU_CTEI. https://t.co/oDcdkwPsif pic.twitter.com/laOAX4w6wC
— Rajiv McCoy (@rajivmccoy) May 25, 2023
Quantitative Biology
For three years, I TA'd the biology department's one-week Quantitative Biology Bootcamp (AS.020.607). In 2020, I also TA'd the fall semester Quantitative Biology Lab course (AS.020.617).
These two courses introduce first-year PhD students — most of whom have no computational experience — to coding and computational biology. They learn Python and bash, and apply their skills to essential biological analyses like sequence alignment, GWAS, and RNA-seq.
In the first year of the pandemic, I also helped convert Quant Bio Bootcamp & Lab into an online/hybrid format for remote learning. This was especially challenging because these hands-on, coding-based courses normally rely on in-person interaction to debug and explain concepts.
In reviews from all years that I TA'd Quant Bio, students repeatedly highlighted the TAs as an integral part of the course's success.
Other teaching experience
Teaching Assistant, Biology REU Data Science Workshop
- 2024: Summer workshop introducing JHU biology REU students to data science in R
Guest Lecturer, Human Genome Variation (AS.020.319)
- 2024: Measuring linkage disequilibrium
- 2023: Quantifying gene expression
- 2023: Data analysis in R
- 2022: Archaic hominin genomics
- 2021: Selection on human structural variants
- 2019: The Wright-Fisher model and population modeling in SLiM
Tutor, Quantiative Biology Lab (AS.020.617)
- 2021-2023: One-on-one tutoring for computational biology assignments
Guest Speaker, Population Genetics Simulation and Visualization (AS.360.111)
- 2023: Quantifying selection with simulation
Teaching Assistant, Developmental Genetics Lab (AS.020.340)
- Fall 2019, Spring 2020: Led students through designing and generating a CRISPR strain in C. elegans